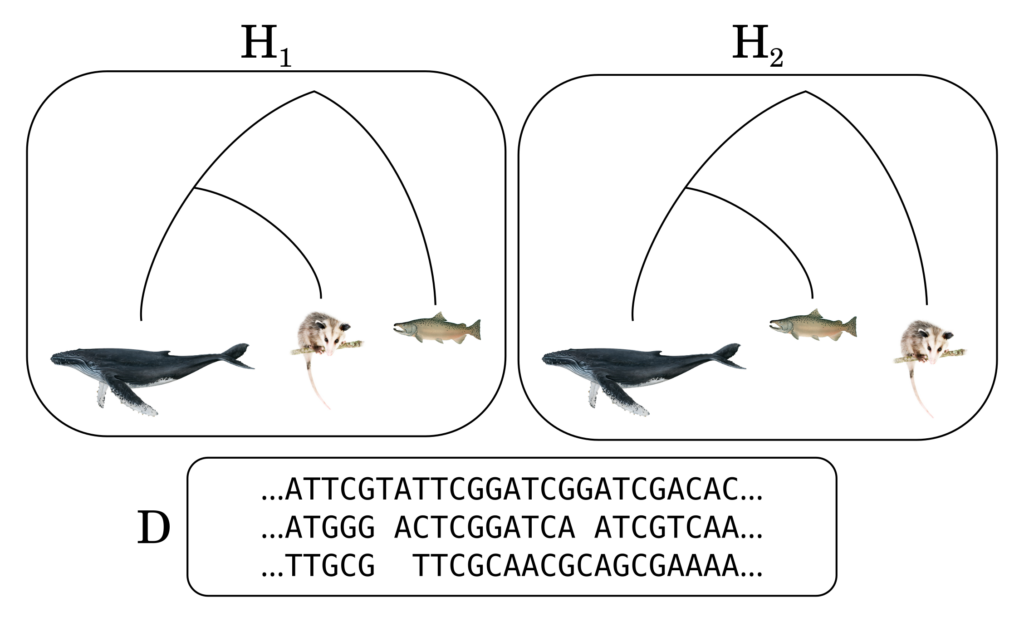
Imagine a conversation with one of these newly released AI chatbots. You ask it it to solve a tricky math problem. It responds with “That seems kind of hard. Give me some time to think.”. After a few minutes it comes back with “I haven’t solve it yet. And I am not sure I can. Would you like me to continue working on it?”. Another few minutes pass and then it comes back with “Aha! I figured it out!” and it proceeds to explain a neat and creative solution.
This scenario can never occur with PaLM, BARD, GPT-4, or any of the other transformer-based large language models that are thought to be on the path to general intelligence. In all of these models, each word in the machine’s response is produced in a fixed amount of time. The model cannot go away and “think” for a while. This is one of the reasons why I believe a solely transformer-based model can never be “intelligent”. (If you disagree with my characterization of transformers here, see section 4 and also this post).
Summary: I argue here, that intelligence requires the ability to explore “trains of thought” that are potentially never-ending. One cannot know a priori if a certain train of thought will lead to a solution or if it is futile. The only way to find out is to actually explore. And this type of exploration comes with the risk of never knowing if you are on the path to a solution or if your current path will go on forever. Intelligence involves problem-solving, and problem-solving requires arbitrary amounts of time. If a computer program is bound to finish quickly by virtue of its architecture, it cannot possibly be capable of general problem-solving.
In the summary paragraph above, I appealed to a number of intuitive notions (e.g. “train of thought”, or “exploration”, or “problem-solving”). In order to make my argument rigorous, I have to first introduce a few concepts rooted in classical theory of computation. In section 1, I will introduce three types of computer programs. In section 2, I describe what an unintelligent problem-solver can look like. In section 3, I describe what is needed to make the unintelligent problem-solver intelligent. In section 4, I explain why transformers can never be general problem-solvers. In section 5, I briefly discuss what I think needs to be done to address this problem.